the idea of reverse complement of a DNA strand is quite easy to understand, although some people spend some time to get around it. Simply put, the procedure can be divided into two main parts. the first and foremost is the process of making a reverse of the given DNA strand.
the second part is making the complement strand of the existing reverse_ strand as shown in the following figure.
an easy algorithm , to make reverse complement, in python 3 would include the following steps.
1- asking user input
2- reversing user input, and printing out the reversed string
3- complementing each nucleotide in a for loop
4- printing out the reverse-complement strand.
below is the syntax of writing the reverse complement script in python 3.5
DNA = input('please enter your DNA sequence below: \n') # asking user input
UpperCase_DNA = DNA.upper() # this line dictates the entry and makes the entry UPPER CASE
print ('your DNA entry is : \n', UpperCase_DNA) # prints out the upper case entry
reverse_DNA = UpperCase_DNA[::-1] # this line reverses the upper case entry
print ("the reverse of your DNA sequence is:", reverse_DNA) # shows the reversed entry [upper cased]
print ('the reverse complement of your entry sequence is: \n') # information
for i in reverse_DNA :
if (i == "A"): print ("T", end = "") # conditional statement to complement each adenine to thymine.
elif (i == "T"): print("A" , end = "")
elif (i == "C"): print("G" , end = "")
elif (i == "G"): print("C", end = "")
I made a video tutorial on this method, i wish it helps somehow ^_^ below is the link to the video .
Click Here
the second part is making the complement strand of the existing reverse_ strand as shown in the following figure.
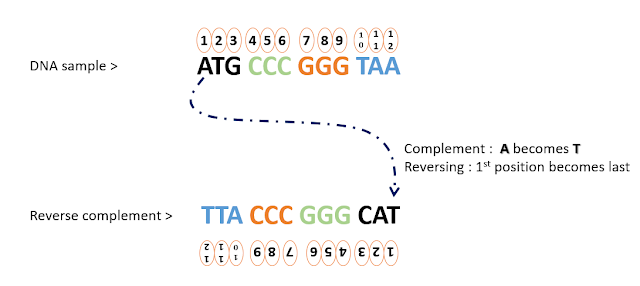 |
| figure 1: process of making reverse complement of a given DNA strand |
an easy algorithm , to make reverse complement, in python 3 would include the following steps.
1- asking user input
2- reversing user input, and printing out the reversed string
3- complementing each nucleotide in a for loop
4- printing out the reverse-complement strand.
below is the syntax of writing the reverse complement script in python 3.5
DNA = input('please enter your DNA sequence below: \n') # asking user input
UpperCase_DNA = DNA.upper() # this line dictates the entry and makes the entry UPPER CASE
print ('your DNA entry is : \n', UpperCase_DNA) # prints out the upper case entry
reverse_DNA = UpperCase_DNA[::-1] # this line reverses the upper case entry
print ("the reverse of your DNA sequence is:", reverse_DNA) # shows the reversed entry [upper cased]
print ('the reverse complement of your entry sequence is: \n') # information
for i in reverse_DNA :
if (i == "A"): print ("T", end = "") # conditional statement to complement each adenine to thymine.
elif (i == "T"): print("A" , end = "")
elif (i == "C"): print("G" , end = "")
elif (i == "G"): print("C", end = "")
I made a video tutorial on this method, i wish it helps somehow ^_^ below is the link to the video .
Click Here

No comments:
Post a Comment